PALEOPARK – Seagrass palaeo-records as a tool for the evaluation, diagnosis and prognosis of the evolution of species, communities, and processes in Spanish insular National Parks
Summary
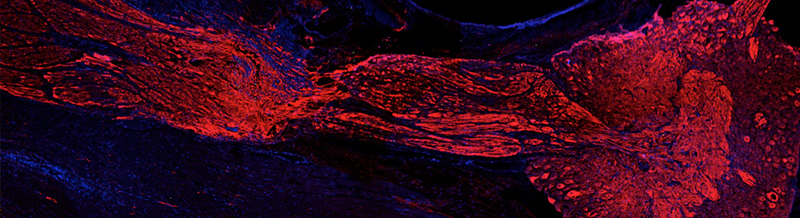
Change in the ecosystems occurs at multiple spatial and temporal scales. Discriminating between real state changes, cycles, and trends, is often difficult or not possible without the adequate time perspective. For this reason, long, detailed, and reliable series of data of relevant ecosystem functional and structural variables, are a central priority for natural reserves managers. Long-term series can be generated from monitoring programs or sought in human, biological, or geological records. Monitoring programs can provide detailed and high-quality information but because of their costly implementation they are often denied by the administrations or drastically limited to a few variables. When the right record is found, modern palaeo-reconstruction techniques can provide an extraordinary wealth of qualitative and quantitative information on environmental and biological features of the ecosystems along very long periods of time and with remarkable time resolution. High human population densities along the coasts of the world, are resulting in constant and intense impact on coastal environments. Paleo-reconstructions can provide valuable insights on these impacts and on the responses of the ecosystems. However, accurate paleo-records in exposed coastal environments are very rare, what has prevented this field of science to develop in such zones. Exceptionally, the endemic Mediterranean seagrass Posidonia oceanica has been found to be an accurate and patient observer of the past natural history of our coasts. Within its highly anoxic peat-like sedimentary deposits, the meadow stores information encompassing, at least, the last ca. 6000 years with a resolution ranging from 1 to 10 yr cm-1. The only other environments where appropriate paleo-records can be found are coastal lagoons. The sheltered condition of these systems, often colonized by species of the genus Zostera, allows the establishment of chronological coherent sedimentary records due to the relatively low hydrodynamism. The peat-like paleo-record formed by P. oceanica, has been found to hold a remarkable stock of carbon in the organic form. In a warming planet, the conservation and enhancement of efficient carbon sinks has become a priority as a help to offsetting the rise in atmospheric CO2. This project proposes 1. to take advantage of these palaeo-records as a tool to characterize the evolution, health state and prognosis, of species, communities, and processes in Spanish insular National Parks (Illas Atlánticas and Cabrera Archipelagos Maritime-Terrestrial National Parks), as driven by humanization and other major environmental stressors and 2. to assess and price the carbon stocks and fluxes associated to the P. oceanica sedimentary record. Both objectives use the same ‘object’ of study and share exactly the same methodology. The objectives will be achieved by the participation of an international consortium of a dozen of research groups that will study geological, chemical, micropaleontological, molecular, genetic, palinological, and isotopic proxies, together with archaeological and historical information. These proxies will provide the managers with information on background levels (pre-anthropic), disambiguation between human and natural derived impacts, identification of ecosystems change patterns (cycles, extreme events, and trends), or quantification of the effects of CO2 rise (sea surface warming and acidification), among many other features.
SZN role
Participant Institution for the analysis of fossil/sub-fossil DNA.
Partners
CSIC, Spain; University of Barcelona, Spain; Universidad Autónoma de Barcelona, Spain; University of Santiago de Compostela, Spain; University Politécnica de Cataluña, Spain; Brunel University, UK; SZN, Italy; University of Western Australia, Australia; NIOZ, The Netherlands; Institut Català d’Arqueologia Clàssic, Spain
Project lifetime
2015-2017
P. I.
Gabriele Procaccini
Project coordinator: Miguel Ángel Mateo Mínguez (CEAB-CSIC, Spain)
Funding Institution
Ministerio de Agricultura, Alimentacion y Medio Ambiente - Spain
RECCAM - Seagrass Meadows resilience to global warming: an analysis based on responses at ecophysiological, population and ecosystem levels
Summary
Climatic change is supposed to cause significant alterations in the global environment, with clear and specific effects in the oceans. The Mediterranean Sea is an excellent model for the study of such effects on marine ecosystems. Seagrass meadows, and specifically those dominated by Posidonia oceanica and Cymodocea nodosa, are amongst the most threatened and relevant Mediterranean habitats. This project is aimed at contributing to the understanding of the main response mechanisms of these key habitats to global warming, probably the main component of climate change.To this end, we have focused the problem through three major approaches, relatively unexplored so far but crucial to achieve a proper knowledge of the impacts of temperature rising. First, we will study the physiological tolerance of Mediterranean seagrass species to thermal stress. Second, we will evaluate the influence of warming on herbivorism. Third, we will analyse the interactions between climatic change and other stressors, in particular eutrophication and mechanical disturbances.
SZN role
Participant Institution for the genetic characterization of seagrass species in mesocosms experiment and for assessment of gene expression in controlled conditions.
Partners
University of Barcelona, Spain; Spanish Oceanographic Institute (IEO), Spain; Stazione Zoologica Anton Dohrn, Italy; CSIC-CEAB, Spain
Project lifetime
2014-2016
P. I.
Gabriele Procaccini
Project coordinator: Javier Romero Martinengo (University of Barcelona, Spain)
Funding Institution
Ministerio de Economía y Competitividad (MINECO) - Spain
Contribution to SZN
€ 16.000
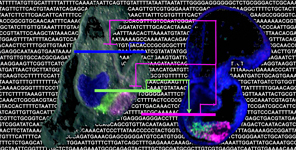
Gut patterning and PANcreas development in evolution and disease: a TRAnsCriptomic approach
Summary
Many genes that have been shown to cause diseases were originally identified because of their role in embryonic development, but were subsequently shown to be also important in the postnatal control of cell growth and differentiation. This is the case of many transcription factors (TF) among which the ParaHox gene Xlox, whose mammalian homolog, Pdx1, is well known for its role in specification of the pancreas, and subsequent formation and maintenance of pancreatic beta-cells. Pdx1 is a causal factor in the development of diabetes, wherein there is a deficiency in insulin production of beta-cells within the pancreas. Moreover, mis-expression of Pdx1 is commonly seen in intestinal disorders such as Crohn’s disease. Here we propose to combine analyses in the highly simple but phylogenetically relevant sea urchin embryo and sea star embryo models with developmentally targeted mouse transcriptome data to characterize regulatory connections that are downstream of the disease-related Xlox/Pdx1 transcription factor.
What we do
We are coordinator of the project and Operating unit SZN and will perform all manipulations and analyses in sea urchin and sea star embryos and all bioinformatic analyses and evolutionary comparisons.
Partners
Stazione Zoologica Anton Dohrn, Napoli; Laboratorio di Medicina Molecolare e Genomica, Università degli Studi di Salerno.
Research Area
Organismal Biology
Project Lifetime
April 2014 to December 2015
SZN Role
Coordinator
Principal Investigator
Funding Institution
MIUR Progetti Premiali (DLGS 213/99)
Contribution to SZN
€169.143,00 (MIUR contribution)
Publications
Annunziata R and Arnone MI (2014). A dynamic network of regulatory interactions explains ParaHox gene control of gut patterning in the sea urchin embryo. Development 141: 2462-72.
Perillo M, Wang YJ, Leach SD and Arnone MI. Specification and differentiation of pancreatic, acinar-like cells in the sea urchin embryo and larva. Submitted to Development.
Meet the team
Maria I. Arnone, primo ricercatore
Rossella Annunziata, postdoc
Claudia Cuomo, PhD student
Elijah Lowe, postdoc

Multidisciplinarity training in evo-devo and neurobiology of marine animal models
Summary
Neptune is a multidisciplinary training network in evo-devo and neurobiology of marine animal models. Through the use of advanced methods of genetic analysis and imaging technologies, Neptune aims at solving an array of important questions in the evolution, development, neurobiology and ecology of marine invertebrates.
Neptune is training a new generation of young researchers by combining the strengths of modern technologies with a real understanding of traditional approaches. The Neptune consortium involves seven academic institutions and one industrial partner that will provide Neptune fellows with expertise, specialized equipment and training on a wide range of approaches and methodologies incorporated in evolutionary developmental biology and marine neurobiology.
What we do
We are one of the seven partners and are contributing to the WP "Evolution of sensory systems" by studying photoreceptor evolution in echinoderms and hemichordates (Ambulacraria).
Partners
European Molecular Biology Laboratory, Heidelberg - DE; Stazione Zoologica Anton Dohrn, Napoli – IT; Uppsala University, Uppsala – SE; Max Plank Institute for Developmental Biology, Tübigen – DE; University College London, London – UK; Sars International Center for Marine Molecular Biology, Bergen – NO; Centre National de la Recherche Scientifique, Villefranches sur mer, Lion - FR; Associate industrial partner: ZEISS.
Research Area
Organismal Biology
Project Lifetime
March 2013 to February 2017
SZN Role
Partner
Principal Investigator
Funding Institution
European Commission, FP7 Call for Proposal: FP7-PEOPLE-2012-ITN
Marie Curie Action - Initial Training Network (ITN)
Grant no. 317172
Contribution to SZN
€302.697,45 (EU contribution)
Dedicated website
Media - Pictures

Publications
Valero-Gracia A, Petrone L, Oliveri P, Nilsson DE, Arnone MI. Non-directional Photoreceptors in the Pluteus of Strongylocentrotus purpuratus Frontiers in Ecology and Evolution (2016) 4, 127.
D’Aniello S, Delroisse J, Valero-Gracia A, Lowe EK, Byrne M, Cannon JT, Halanych KM, Elphick MR, Mallefet J, Kaul-Strehlow S, Lowe CJ, Flammang P, Ullrich-Lüter E, Wanninger A and Arnone MI (2015). Opsin evolution in the Ambulacraria. Marine Genomics, 24: 177-183.
Ullrich-Lüter E, D’Aniello S and Arnone MI (2013). C-opsin expressing photoreceptors in echinoderms. Integr Comp Biol 53: 27-38.
Peterson KJ, Su Y-H, Arnone MI, Swalla B, and King B (2013). microRNAs Support the Monophyly of Enteropneust Hemichordates. J Exp Zool B Mol Dev Evol 320: 368-374.
Meet the team
Maria I. Arnone, primo ricercatore
Alberto Valero Gracia, PhD student
Marie Curie Career Integration Grant (FP7-PEOPLE-2011-CIG)
Summary
The main interest of the project is to study the Nitric Oxide Synthase (NOS) family evolution and its regulation during amphioxus development. In the framework of the project, setting-up an amphioxus facility at the SZN was a priority of national interest, representing the first Italian laboratory working with live amphioxus embryos on demand.
Since Furchgott, Ignarro and Murad won the Nobel Prize in Medicine or Physiology in 1998 for their breakthrough work on the role of nitric oxide (NO) as a multifunctional signaling molecule, many reports have shown the seemingly limitless range of body functions controlled by this compound. To manipulate the endogenous NO level for therapeutic benefits using NOS gene therapy is essential to understand the physiological and developmental functions of different NOS isoforms (nNOS, iNOS, eNOS). Due to their extensive conservation over evolutionary time, one would expect greater differences and structural changes in NOS genes than that we have observed (Andreakis 2011), reflecting the very ancient and essential nature of Nitric Oxide biological pathways.
Surprisingly, a single molecule, identical in all living animals, can fulfil a huge range of different functions. This suggests that differences in the regulation of NOS enzymes expression are key in explaining their functional diversification, functional novelties and degree of complexity.
What we do
We use as animal model system the cephalochordate amphioxus Branchiostoma lanceolatum, from the Gulf of Napoli (Italy) and from Banyuls-sur-Mer (France), with comparative and multidisciplinary approaches in the field of Evolutionary and Developmental Biology (Evo-Devo). The primary aim of this project is to perform a detailed study of the duplicated set of NOS genes during amphioxus development, trying to establish the basic primary NOS roles that are evolutionary conserved in chordates.
Partners
Stazione Zoologica Anton Dohrn, Napoli.
Research Area
Organismal Biology
Project Lifetime
August 2011 to July 2015
SZN Role
Coordinator
Principal Investigator
Funding Institution
European Commission, FP7 Call for Proposal: FP7-PEOPLE-2011-CIG
Grant no. 293871
Contribution to SZN
€100.000 (EU contribution)
Dedicated website
http://cordis.europa.eu/project/rcn/99685_en.html
Publications
Coppola U, Annona G, D’Aniello S* and Ristoratore F* (2015). Rab32/38 duplicated genes in chordate pigmentation: an evolutionary perspective BMC Evolutionary Biology, under review.
Annona G, Holland ND* and D’Aniello S* (2015). Evolution of the notochord. EvoDevo, in press.
Anishchenko E and D’Aniello S* (2015). Tunicate neurogenesis: the case of the SoxB2 missing CNE. Mathematical Models in Biology (Springer), in press.
Vassalli QA, Anishchenko E, Caputi L, Sordino P, D'Aniello S* and Locascio A* (2015). Regulatory elements retained during chordate evolution: Coming across tunicates. Genesis 53: 66-81.
Pascual-Anaya J, D’Aniello S, Kuratani S and Garcia-Fernàndez J (2013). Evolution of the Hox clusters in deuterostomes. BMC Developmental Biology 13: 26.
Meet the team
Salvatore D’Aniello, Ricercatore
Evgeniya Anishchenko, post-doc
Giovanni Annona, post-doc
Filomena Caccavale, PhD student